ChemBioML Platform enables easy and reproducible machine learning for chemical and biological data with the extensive documentation. It supports training Ridge regression models using Genetic Algorithms and making predictions from trained models.
Input File Format
Format of Input Training File is same for all versions of ChemBioML Platform:
Format: .xlsx file
Required sheets: “Train” and “Val”
Each sheet must contain:
The endpoint column (target value)
Independent variables (features)
The same column structure in both sheets
Example file is available here.
ChemBioML GUI Documentation
Grafical version of ChemBioML Platform links powerful capabilities of Unreal Engine 5 together with Python`s ability to train accurate and robust Machine Learning models.
Installation
Download and unpack ZIP file with ChemBioML Platform to any folder on your PC – and it`s ready to work! Use ChemBioML Launcher.vbs to run the software.
Select the task

In Grafical version documentation can be accessed via meny “Help”.
Train
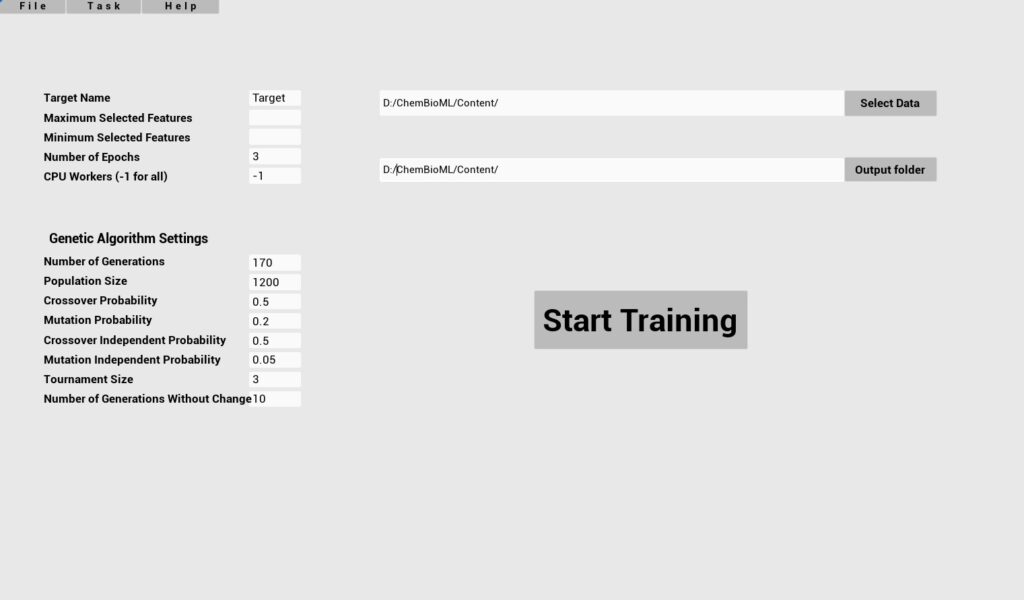
Required parameters
Key
Select Data
Target Name
Maximum selected features
Minimum selected features
Number of Epochs
Output Folder
Description
Full path to .xlsx file with training data
Name of the target column
Maximum number of features
Minimum number of features
Number of training epochs
Path to save model
Optional parameters
Key
CPU Workers
Population Size
Crossovers probability
Mutation probability
Number of generations
Crossover independent probability
Mutation independent probability
Tournament size
Number of generations no change
Default
-1
1200
0.5
0.2
170
0.5
0.05
3
10
Predict
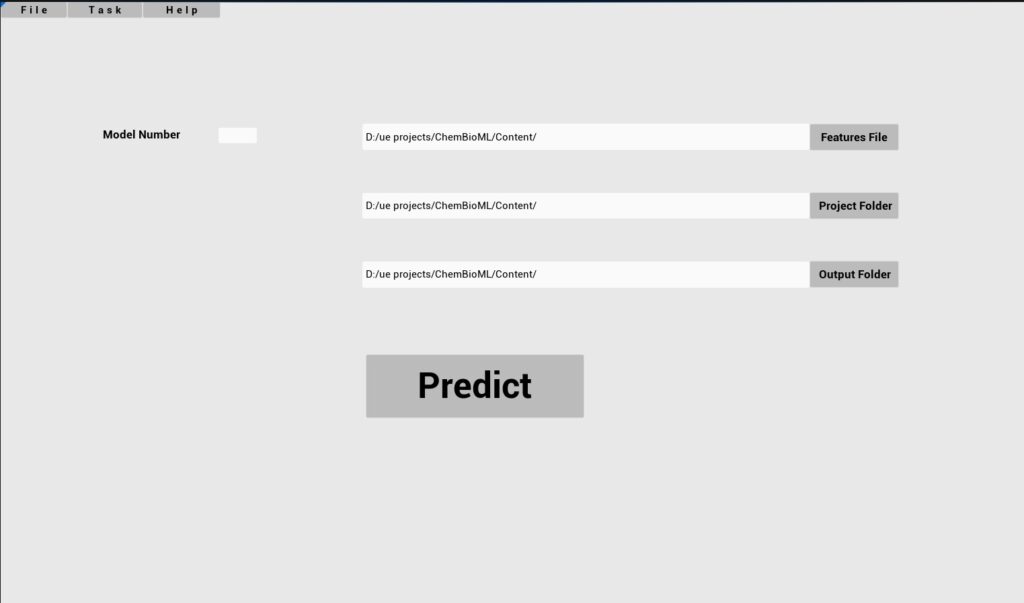
Required parameters
Key
Features file
Project Folder
Model Number
Output folder
Description
Path to file with input features
Folder where the model is stored
ID of the trained model to use
Output folder Path to save predictions
ChemBioML CLI Documentation
The embedded version of ChemBioML allows users to train or predict using a simple config.txt file and a launcher script (ChemBioMLauncher.bat). No command-line knowledge is required.
Installation
Same as GUI version!:)
Configuration File Format (config.txt)
Use this file to control what the software does. All required parameters must be set correctly before launch.
Choose Command
Enable only one of these two lines:
command=Train_linear_Ridge_GA_Regressor
# command=Predict_linear_Ridge
Train a Ridge Model (Train_linear_Ridge_GA_Regressor)
Required parameters
Key
input_path
endpoint_name
max_features
min_features
n_epochs
output_path
Description
Full path to .xlsx file with training data
Name of the target column
Maximum number of features
Minimum number of features
Number of training epochs
Path to save model and logs (applies to both commands)
Optional parameters
Key
n_cpu
n_population
crossover_proba
mutation_proba
n_generations
crossover_independent_proba
mutation_independent_proba
tournament_size
n_gen_no_change
Default
-1
1200
0.5
0.2
170
0.5
0.05
3
10
Description
Number of CPU cores to use
GA population size
Crossover probability
Mutation probability
Number of generations
Independent crossover probability
Independent mutation probability
Tournament selection size
Stop after N unchanged generations
Example section
command=Train_linear_Ridge_GA_Regressor
input_path=”C:/…/se_for_tML.xlsx”
endpoint_name=IC50
max_features=4
min_features=1
n_epochs=1
output_path=”C:/…/models”
Predict with a Trained Model (Predict_linear_Ridge)
Required parameters
Key
features_for_prediction
project_path
model_number
output_path
Description
Path to file with input features
Folder where the model is stored
ID of the trained model to use
Path to save predictions
Example Section:
command=Predict_linear_Ridge
features_for_prediction=predict_data.xlsx
project_path=model_dir/
model_number=1
output_path=”C:/…/predictions”
How to Run
After editing config.txt, launch the software using:
ChemBioMLauncher.bat
ChemBioML OS Documentation
The Open-Source version of ChemBioML Platform enables easy and reproducible machine learning for chemical and biological data. It supports training Ridge regression models using Genetic Algorithms and making predictions from trained models.
Installation
Install ChemBioML via pip:
pip install -U chembioml
After installation, use it directly via command line:
chembioml <command> [arguments]
Available Commands
Train_linear_Ridge_GA_Regressor
Train a Ridge Regression model using a Genetic Algorithm.
Required Arguments:
Argument
–input_path
–endpoint_name
–max_features
–min_features
–output_path
Description
Path to .xlsx file with training data
Name of the target (endpoint) column
Max number of features to use for training
Min number of features to use for training
Directory to save the trained models and logs
Optional Arguments:
Argument
–n_epochs
–n_cpu
–n_population
–crossover_proba
–mutation_proba
–n_generations
–crossover_independent_proba
–mutation_independent_proba
–tournament_size
–n_gen_no_change
Default
3
-1
1200
0.5
0.2
170
0.5
0.05
3
10
Description
Number of training epochs
CPU cores to use (-1 = all)
GA population size
Probability of crossover
Probability of mutation
Number of generations
Independent crossover probability
Independent mutation probability
Stop after N unchanged generations
Example
Unix system:
chembioml Train_linear_Ridge_GA_Regressor \
–input_path data.xlsx \
–endpoint_name IC50 \
–max_features 50 \
–min_features 10 \
–output_path ./trained_models
Windows system:
chembioml Train_linear_Ridge_GA_Regressor ^
–input_path data.xlsx ^
–endpoint_name IC50 ^
–max_features 50 ^
–min_features 10 ^
–output_path ./trained_models
Predict_linear_Ridge
Use a trained Ridge model to make predictions on new feature data.
required arduments
Argument
–features_for_prediction
–project_path
–model_number
–output_path
Description
Path to .xlsx file with features only
Path to the folder with trained models
Model ID to use for prediction
Path to save the prediction results
example
Unix system:
chembioml Predict_linear_Ridge \
–features_for_prediction new_data.xlsx \
–project_path ./trained_models \
–model_number 2 \
–output_path predictions
Windows system:
chembioml Predict_linear_Ridge ^
–features_for_prediction new_data.xlsx ^
–project_path ./trained_models ^
–model_number 2 ^
–output_path predictions
For CLI and OS versions Documentation can be accessed only within this page.